Resources
|
Computational Tools
Code for several of these tools is freely available on SourceForge or Github. |
Review Articles and Protocols
|

An open-source Python tool which provides a common API to native file formats from multiple mass spectrometry manufacturers.
mzAPI: a new strategy for efficiently sharing mass spectrometry data. Nat Methods 2009;6:240-1. 
A Python desktop application that combines our mzAPI with other proteomic-centric tools.
Multiplierz v2.0: a python-based ecosystem for shared access and analysis of native mass spectrometry data. Proteomics 2017;17:1700091. 
An interntet application that enables researchers to easily transition from peptides identified by mass spectrometry to biological pathways.
Pathway Palette: A rich internet application for peptide-, protein- and network-oriented analysis of MS data. Proteomics 2010;10:1880-85. 
An interactive viewer that enables efficient distribution of proteomic results and analysis of the native mass spectrometry data files .
mzresults: an interactive viewer for interrogation and distribution of proteomics results. Mol Cell Proteomics 2011;10:M110.003970, 1-7. 
A web-based server that exposes our mzAPI through intuitive RESTful uniform resource locators to provide remote data access and analysis capabilities.
mzserver: web-based programmatic access for mass spectrometry data analysis. Mol Cell Proteomics 2011;10:M110.003988, 1-7. 
An open-source framework that enables users to generate arbitrarily complex LC-MSn acquistion methods through simple Python scripting.
Library dependent lc-ms/ms acquisition via mzapi/live. Proteomics 2013;13:1412-16. |
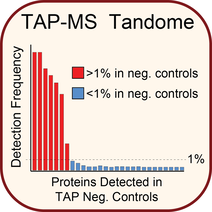
A detailed step-by-step protocol of our approach for tandem affinity purification combined with mass spectrometry.
Tandem affinity purification and mass spectrometry (TAP-MS) for the analysis of protein complexes. Curr Prot Protein Sci 2019; in press. 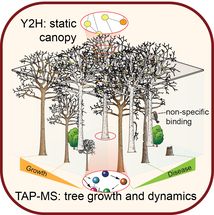
A discussion of the advantages of studing proteins complexes formed under physiologic conditions.
Protein complexes: the forest and the trees. Expert Rev Proteomics. 2009;6:5-10. 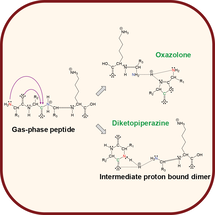
A review of multiple topics in modern proteomics with a useful primer for manual interpretation of MS/MS spectra.
Mass spectrometry-based proteomics: qualitative ID to activity-based protein profiling. Wiley Systems Biology and Medicine 2012;4:141-62. |
HOME | RESEARCH | PEOPLE | PUBLICATIONS | RESOURCES | CONTACT
©2023 Marto Laboratory at Dana-Farber Cancer Institute
©2023 Marto Laboratory at Dana-Farber Cancer Institute
